Genes of bacteria which have a high G+C content genome DNA such as
Streptomyces possess biased codon usage. This results in
extremely high G+C distribution at the third-letter of each
codon. Streptomyces genes actually have an average third-letter
G+C content of 92%, which was calculated from 2,008
Streptomyces genes recorded in the CUTG database (Nakamura et al., 1998) based on the
GenBank database release 108. This characteristic enables the
prediction of protein-coding region in such bacteria. The Frame
analysis was first developed by Bibb et al. (1984), and it is one of the essential analyses
for studying Streptomyces genetics even at the present time
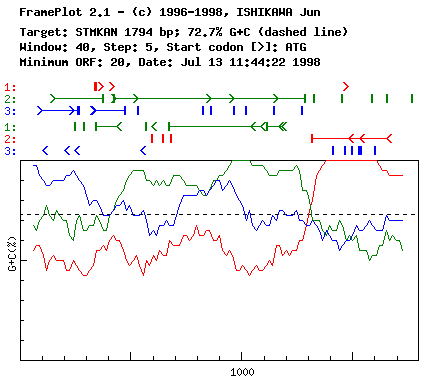
Sample Results
Main page
-
DNA sequence:
- Enter your DNA sequence.
Don't include comments.
Digits and spaces are ignored.
The program substitutes T for U.
-
Window size (default = 40 codons):
- Choose the size of sliding window.
-
Start codon (default = ATG):
- Choose the codon(s) recognized as start codon.
-
Step Size (default = 5 codons):
- Window is moved along the sequence by
the chosen step size.
Larger step size makes the program run much faster.
It will produce low resolution plot,
but using of the default value gives a satisfactory result
in nine cases out of ten.
-
Minimum ORF size (default = 20 codons):
- The program recognizes ORFs which are larger than
minimum ORF size as ORF.
-
Incomplete ORF (default ON)
- The program recognizes incomplete ORFs which don't have
apparent start or stop codon.
-
Image width (default = auto):
- Choose the width of the output image.
By default, Image width is adjusted automatically.
-
Image color (default = color):
- Choose the color of the output image (color or black/white).
-
Sequence label:
- Specify a description for the sequence.
This does not affect the result.
Result page
- Clickable Map:
- You can get nucleotide and amino acid sequence (FASTA format)
of the ORF you interested by clicking on the graph.
Translation is performed as following manner:
1 2 3 E
>---100aa--->--50aa-->-10aa-|
A B C
- clicking on A yields translation from the start codon 1.
- clicking on B yields translation from the start codon 2.
- clicking on C yields translation from the start codon 2 too.
That is because the ORF (3 to E) is smaller, by default,
than minimum ORF size.
Features page
- BLAST Gateway:
- You can submit the sequence immediately to the NCBI BLAST server
by clicking of [Submit] button.
References
-
Bibb, M.J., Findlay, P.R. and Johnson, M.W. (1984)
- The relationship between
base composition and codonusage in bacterial genes
and its use for the simple and reliable identification
of protein-coding sequences.
Gene 30: 157-166.
-
Ishikawa, J. and Hotta, K. (1991)
- Nucleotide sequence and transcriptional start point of
the kan gene encoding
an aminoglycoside 3-N-acetyltransferase
from Streptomyces griseus SS-1198PR.
Gene 108: 127-132.
-
Ishikawa, J. and Hotta, K. (1999)
- FramePlot: a new implementation of the Frame analysis
for predicting protein-coding regions in bacterial DNA with
a high G+C content. FEMS Microbiol. Lett. 174:251-253.
-
Nakamura, Y., Gojobori, T. and Ikemura, T. (1998)
- Codon usage tabulated from the international DNA sequence databases.
Nucl. Acids Res. 26:334.
